On January 23, 2018, this June 19, 2014 post was reconstructed due to new information about the families discussed herein. When it was originally published, our three surname researchers had been under the estimation for nearly three decades that the Owston family from Ganton in the original East Riding of Yorkshire was originally descended from Giles Owston (d. 1641) and his progeny who lived in this same parish from the 16th through the 18th centuries.
However, recently released Big Y results and a reexamination of the records of three parishes indicate that the current Ganton Owstons are actually a branch of the Sherburn family and not a distinct family at all. This was something that was not overtly evident in the original records and could not be determined with either STR or autosomal DNA testing.
While the new data places these participants at a closer relationship level than originally thought, it does not conflict with our original premise from 2014. In addition, more recent results are used in this update, as we are able to move the comparison from 59 to 253 relationships.
INTRODUCTION:
While obviously having a small pool of potential Y-DNA participants, low frequency surnames may have the advantage of having good documentation of ancestry. That is the case with my surname and its corresponding Y-DNA Project. The original intent of our project was to see if three families (at the time) bearing our surname from the original East Riding of Yorkshire had a common ancestry or if the surname was applied to these lineages independently from each other. Since the original writing, the number of distinct families has been reduced from three to two.
The two extant families are as follows:
| SHERBURN: The largest Owston family hails originally from Sherburn in Harford Lythe and its many members descend from Peter Owston who died in 1568. This group also includes the Ouston descendants of James Ouston (1711-1785) who was born in Brompton by Sawdon and who died in Sigglesthorne. You’ll find members of this clan in the UK, Australia, USA, Canada, New Zealand, Nicaragua, and Italy. Ninety-one percent (yes, we counted) of all Owston and Ouston males descend from the Sherburn family. |
| THORNHOLME: The second group of Owstons from 15 miles south of Sherburn and Ganton can be traced to Richard Owston of the village of Thornholme in the parish of Burton Agnes. Richard Owston died in 1739. By using onomastic evidence, it is possible to theorize a connection to an earlier Ganton line fathered by Robert Owston who was born in the 16th century. The names Richard, Michael, and Matthew appear both in the later Thornholme family as well as in the extinct Robert Owston of Ganton family. The Thornholme Owstons are found in Canada, Australia, the UK, Finland, and New Zealand. Only 25 males descend from the smaller Thornholme family. |
In the study’s first year, a positive conclusion was reached; as three participants (one from each identified family at the time) matched each other at 100% using 43 Y-DNA marker tests from GeneTree. Others in the study matched at a genetic distance of 2 and 3.
This was exciting news, as it was impossible to determine a relationship between these lineages, as the connection between these families apparently occurred before the introduction of English parish registers in 1538.
The first record of the surname in the region (spelled as Oustyn) appeared in a 1452 will from the parish of Wintringham. By the 16th century, the surname began to ramify within a five mile radius of Ganton and was found in Ganton, Staxton in Willerby, Sherburn, Foxholes, and West Heslerton - all presently in the Ryedale District of North Yorkshire.
GROWTH OF THE PROJECT:
In order to better understand our relationships and to construct a more conclusive modal haplotype of the Owston families, it was necessary to branch out beyond our original participants and attempt to test as many Owston/Ouston males as possible. We have identified 23 lines from the two families. Some lines can further be subdivided into groups that we call segments. Currently, there are 37 segments and unsegmented lines.
As of 2018, the Owston/Ouston DNA project has 33 Y-DNA participants – 28 Sherburn family members and 5 Thornholme participants. The participants represent at least one person from 20 of the 23 lines and 26 of the 37 lines and segments. Additionally, some lines/segments have more than one participant. All five Thornholme lines are represented. For a look at the charts, see http://www.owston.com/family/owston/Owston_Family_Charts.pdf.
Of the 33 participants, one individual is awaiting awaiting test results. Nine of the remaining individuals failed to match the modal haplotype and are apparently the results of non paternal events (NPEs). Several of these participants descended from families where known NPEs existed, while others’ results were a complete surprise. It is to be noted that everyone who tested had a clear paper genealogical line to one of the two original families. It is estimated that one-third of Owston/Ouston males have an ancestral NPE; this number may be even greater.
GENETIC DISTANCE:
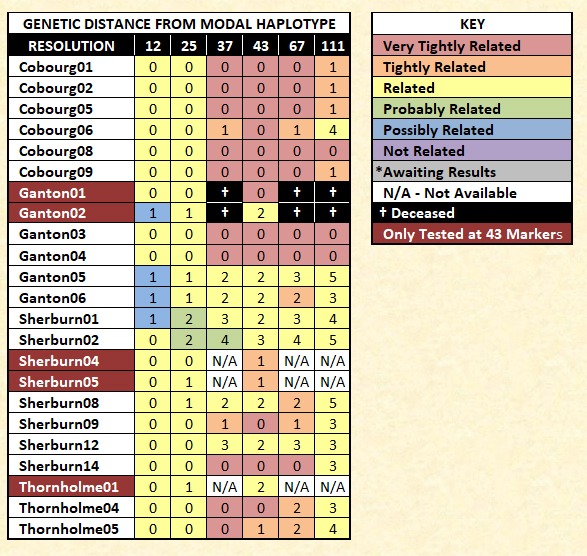
Currently, 23 of the participants have a solid match to the modal haplotype. These represent three Thornholme participants and 20 Sherburn participants. Early in this study, we noticed that some individuals were inclined to have closer genetic matches with participants who were genealogically more distant than those who were more closely related. This was a curiosity that led to the eventual writing of this post and the raising of 18 participants' STR resolutions to 111 markers. It also influenced our testing of 15 participants with FTDNA's Big Y test (3 tests are currently pending).
Over the years, genetic genealogists have tended to rely upon genetic distance to help predict a range of possible relationships. In fact, FTDNA qualifies matches at various levels of genetic distance.
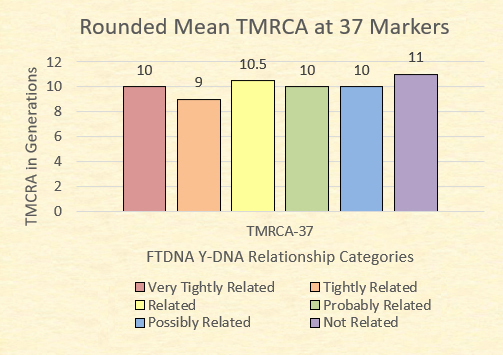
For example, FTDNA states that a GD=0 at 37 markers indicates that the two individuals are “very tightly related”; and with a confidence level of .05 or less, FTDNA estimates that these individuals are related within eight generations (seventh cousins). A mismatch of one GD is considered “tightly related.” Genetic distances at 2 or 3 marker differences between men of the same surname are identified as “related.” As GD increases, the likelihood of a relationship diminishes with a GD=6 as considered as being not related, even when the same surname is present (Canada, 2011).
In addition, most of us have genetically close matches with individuals who obviously are further back on the relationship continuum and do not share a common surname or variant. At 37 markers, I have a number of matches with individuals whose ancestry hails from distances far removed from my own East Riding ancestors. While we are related, the connection is likely before the genealogical time frame and may be prior to the various invasions of Britain – one of which brought my ancestors from mainland Europe.
Using my project as a case study, I have hypothesized that, although a predictor of a familial connection, genetic distance is an inadequate predictor of relationships. Before I discuss my results, I must present some caveats.
First of all, I cannot confirm the exact connection between the two families in my study; however, I have constructed plausible trees based on shared forenames, typical naming conventions, names found in wills and other local records, and the close geographical distances among both current families and three earlier extinct families. Currently, we can only affirm the relationship intra-family; however, based on the aforementioned factors, we are confident that the supposed relationships are close to the unknown actual relationships. The closest possible relationships are used in this analysis and should be no more distant than two generations, but due to different downstream SNP markers, the relationships are not any closer.
Secondly, not all 23 matching participants tested at the same level of resolution. Five participants tested only at 43 markers at the now defunct GeneTree. Two of these men are deceased. We have been attempting to get the remaining three to upgrade at Family Tree DNA. Hopefully, at least two key participants will agree to a retest in 2018. One is critical as he is the last male in his line. We were successful in 2016 in retesting the last male in another line, as he died in late 2017.
In the original post, we compared all 37 marker tests to each other and all 43 marker tests to each other. The original combined analysis had only 59 compared relationships. In this new analysis, we compare 153 relationships at 37 markers and 253 at 43 markers. We only include the 43 marker results in order to use the data from the five participants who have only tested at 43 markers. These five can be compared to the 18 participants who have tested at 111 markers, as the additional markers at 43 are found within FTDNA's 111 STR markers.
Third, there were only five participants with close relationships (fifth cousins and closer) in the original analysis; therefore, the mean TMRCA (time to the most recent common ancestor) skewed more distant. We now have 26 relationships at the fifth cousin level or closer.
The relationships are as follows:
| Relationships | Number |
| Father/Son | 1 |
| Brothers | 1 |
| Uncle/Nephew | 2 |
| 1st Cousins, Once Removed | 1 |
| 2nd Cousins | 3 |
| 2nd Cousins, Once Removed | 3 |
| 3rd Cousins, Once Removed | 1 |
| 4th Cousins | 7 |
| 4th Cousins, Once Removed | 4 |
| 5th Cousins | 3 |
| 5th Cousins, Once Removed | 5 |
| 6th Cousins | 2 |
| 6th Cousins, Once Removed | 1 |
| 7th Cousins | 4 |
| 7th Cousins, Once Removed | 6 |
| 7th Cousins, Twice Removed | 1 |
| 8th Cousins | 23 |
| 8th Cousins, Once Removed | 29 |
| 8th Cousins, Twice Removed | 4 |
| 9th Cousins | 30 |
| 9th Cousins, Once Removed | 21 |
| 9th Cousins, Thrice Removed | 2 |
| 10th Cousins | 4 |
| 10th Cousins, Twice Removed | 24 |
| 11th Cousins, Once Removed | 10 |
| 12th Cousins* | 1 |
| 12th Cousins, Once Removed* | 14 |
| 12th Cousins, Twice Removed* | 5 |
| 12th Cousins, Thrice Removed* | 1 |
| 13th Cousins* | 24 |
| 13th Cousins, Once Removed* | 10 |
| 13th Cousins, Twice Removed* | 5 |
| 13th Cousins, Thrice Removed* | 1 |
| *The exact relationships between members of the Sherburn and Thornholme families are not presently known. These relationships are estimated through onomastic evidence and references in wills; the assumed relationships are listed at the closest possible level; however, they may be one or two more generations further distant, but not any closer, as the families have distinct SNP signatures beyond their common I-A10206 SNP. |
A relationship listed with a .5 value (i.e., 1.5, 5.5, 14.5) indicates that the two parties are from different generations. For instance, 9th cousins, once removed are categorized as having at TMRCA of 10.5 generations. One member of the pair has a TMRCA of 10 generations, while the other individual has a TMRCA of 11 generations. The compromise is 10.5 generations for the relationship's common ancestor.
37 MARKER ANALYSIS:
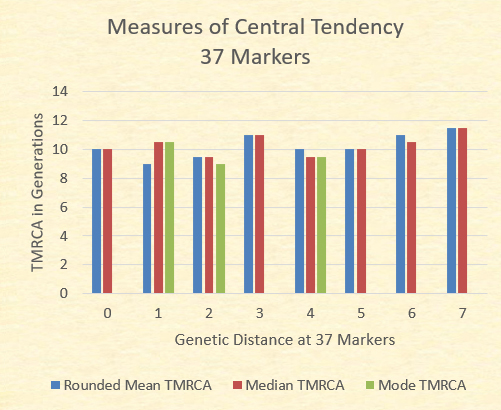
Often considered a baseline reliable STR resolution, the 37 marker test from FTDNA is generally considered the entry point for many Y-DNA participants. Of our 153 relationships at this resolution, the following chart illustrates the range of time to the most recent common ancestors (TMRCA) and the mean (mathematical average). Finally, the mean is rounded to the nearest .5 to determine the adjusted mean relationships.
| GD | n | MIN TMRCA | MAX TMRCA | MEAN TMRCA | ADJUSTED MEAN RELATIONSHIP |
| 0 | 46 | 1.0 | 14.5 | 9.90 | 9th Cousins |
| 1 | 20 | 3.0 | 14.5 | 9.20 | 8th Cousins |
| 2 | 31 | 3.5 | 14.5 | 9.69 | 8th Cousins, Once Removed |
| 3 | 28 | 5.0 | 15.0 | 11.11 | 10th Cousins |
| 4 | 16 | 5.5 | 13.5 | 10.06 | 9th Cousins |
| 5 | 9 | 8.5 | 12.0 | 10.06 | 9th Cousins |
| 6 | 2 | 10.0 | 12.0 | 11.00 | 10th Cousins |
| 7 | 1 | 11.5 | 11.5 | 11.50 | 10th Cousins, Once Removed |
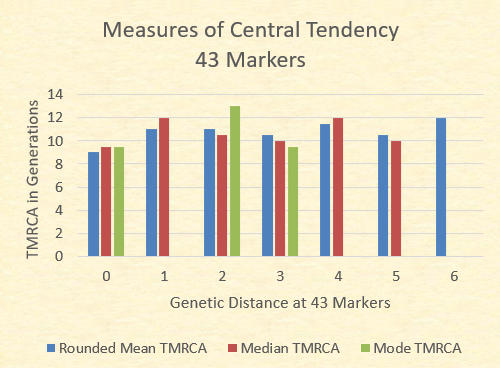
While the mean TMRCA The chart below illustrates the mean (adjusted to the nearest .5), the median (the midpoint TMRCA in the series), and the mode (most common TMRCA value). Where no green bar is present, there are multiple modes.
With the exception of the GD=7 where there is only one sample, the mean TMRCA, median TMRCA, and the modal TMRCA all fall within one generation of 10 generations.
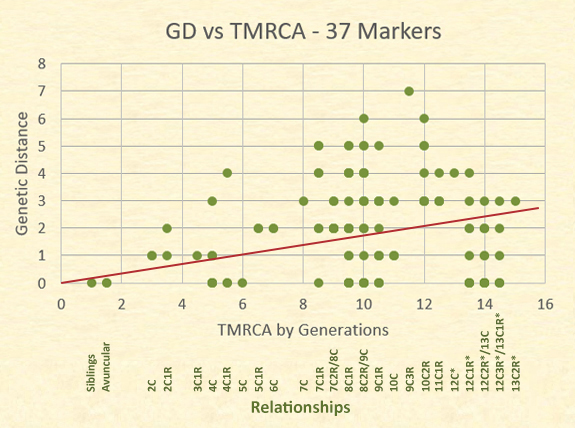
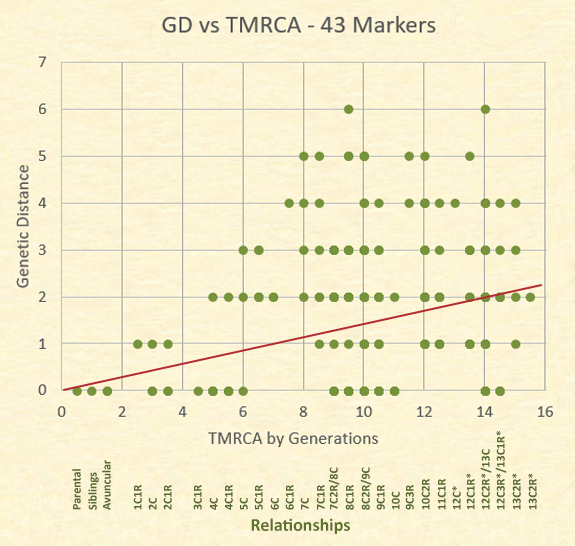
The actual relationships and their genetic distances are better represented, however, on a scatter plot. Although there are 63 dots represented in the plot, there are 153 relationships.
The dots in the following graph often represent several distinct relationships at the same level and genetic distance. For example, the dot for a TMRCA of 14 generations (12C1R/13C) and a GD=0 represents 8 relational pairs; whereas the dot for siblings at a GD=0 represents only one relational pair. The trend line is close to the mid point of the results.
Notice the number of relationships showing a GD=0. Those at the highest relationship level are pairs of individuals who have a common ancestor born in the 1400s, and yet 16 of these distant cousins have a GD=0. More variability, however, is shown in the area of eighth cousins, twice removed and ninth cousins with a genetic distance range of 0 to 6.
Relationships with a GD of 5, 6, and 7 are indicative of each individual of the pair having more STR mutations from the modal haplotype than other typical participants. These differences often become wider at greater testing resolutions as illustrated below.
Although bearing their own designations, both the Cobourg (named for a Canadian town) and the Ganton families are subsets of the Sherburn family.
Finally, FTDNA categorizes relationships at 37 markers as being very tightly related (GD=0), tightly related (GD=1), related (GD=2 or 3), probably related (GD=4), possibly related (GD=5), and not related (GD=6+).
Note, FTDNA will only show matches with a genetic distance of four or less. Twelve relationships do not show as matching at 37 markers. These relationship pairs are as follows:
- one seventh cousin, once removed pair;
- one seventh cousin, twice removed pair;
- two eighth cousin, once removed pairs;
- one eighth cousin, twice removed pair;
- two ninth cousin pairs;
- one ninth cousin, once removed pair;
- one ninth cousin, thrice removed pair;
- and three tenth cousin, twice removed pairs.
43 MARKER ANALYSIS:
I questioned whether I should include the analysis at 43 markers, as no company is presently (to my knowledge) offering 43 (also marketed as 46) marker STR tests. I decided to include the results based on two reasons. First, the original 2014 post used 43 marker tests in its analysis. Second, adding the five participants increases the number of relationship analysis from 153 pairs to 253 pairs. The data for these additional 100 relational pairs could prove valuable; however, the overall analysis will concentrate on the 37 marker data.
The data for 43 markers uses similar tables and charts to those used in the 37 marker analysis.
| GD | n | MIN TMRCA | MAX TMRCA | MEAN TMRCA | ADJUSTED MEAN RELATIONSHIP |
| 0 | 67 | 0.5 | 14.5 | 8.84 | 8th Cousins |
| 1 | 38 | 2.5 | 15.0 | 11.24 | 10th Cousins |
| 2 | 78 | 5.0 | 15.5 | 10.85 | 10th Cousins |
| 3 | 41 | 6.0 | 15.0 | 10.54 | 9th Cousins, Once Removed |
| 4 | 17 | 7.5 | 15.0 | 11.71 | 10th Cousins, Once Removed |
| 5 | 10 | 8.0 | 13.5 | 10.25 | 9th Cousins, Once Removed |
| 6 | 2 | 9.5 | 14.0 | 11.75 | 11th Cousins |
While the data presented at 43 markers is somewhat different, the results still indicate a great variety of relationships found at particular genetic distances. Although a perfect trend would be non-linear, the linear representation roughly indicates the middle of all the results. Remember, each dot can represent multiple relationships.
CONCLUSION:
The average results across the board are not significantly different. At a 37 marker resolution, a GD=0 and a GD=5 result in the same adjusted mean relationship: 9th cousins. The differences are found with the range of results. In our examples, a greater genetic distance indicates a narrower range of possible relationships. Therefore, it is unlikely that two people with a GD=4 are close relatives; however, a GD=0 could represent numerous relationships from very close relatives to those who are very distant, as a genetic distance of zero is all over the road.
In addition, matches with individuals of different surnames also create prediction problems. By analyzing the genetic distance, it is impossible to determine if any of these matches are related within the genealogical time period. In my own match list, I have 67 other surnamed individuals who match me at genetic distances of two, three, and four. Do any share an ancestor within the last 600 years and are a product of an ancestral NPE? Do they share an ancestor from the distant past and their results mimic close relationships due to convergence? At 37 markers alone, it is impossible to tell.
According our results, genetic distance at 37 markers is an insufficient predictor of relationship.
Although we have reached this conclusion, this is just one study and the results may only be indicative of this particular surname. I would be curious in knowing if others can replicate similar results in their studies.
While we may never know what causes the frequency of mutations on Short Tandem Repeats, the examination of studies where ancestries are documented may help us to better understand the role that genetic distance plays and to be able to better understand relationship prediction.
REFERENCE:
Canada, R.A. (2011). If two men share a surname, how should the genetic distance at 37 Y-Chromosome STR markers be interpreted? Family Tree DNA. https://www.familytreedna.com/learn/y-dna-testing/y-str/two-men-share-surname-genetic-distance-37-y-chromosome-str-markers-interpreted/